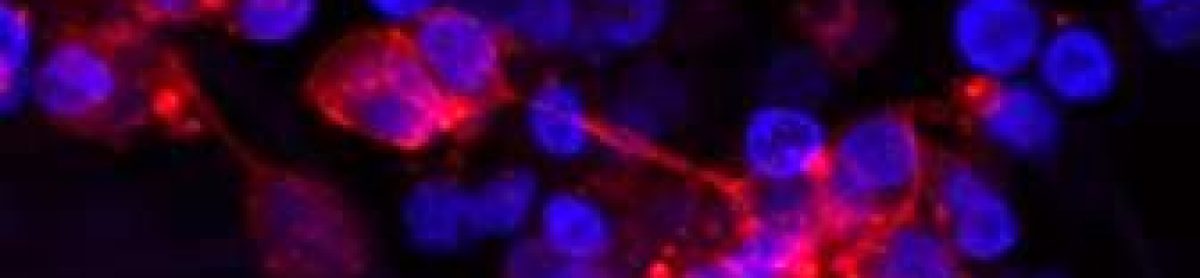
Researchers use various methods, but I employ gene knockdown in my experiments. Basically, I use small RNA molecules that specifically target and degrade the mRNA of my gene of interest. This leads to a decrease in the corresponding protein levels, enabling me to observe the effects on neuroblastoma cell behaviour.
I feel a bit like Sherlock Holmes, you know? I’m selectively putting my suspect protein – the one I’m eyeing – under the spotlight to see how it’s pulling the strings on the cell’s behaviour. It’s like I’m in a cellular mystery, complete with a gene knockout magnifying glass 🔍🧬🕵
So, what I’ve been up to these past months is knocking down my protein and trying to find answers to the following questions:
Can neuroblastoma cells survive? And if not, how do they meet their demise? Do they go on a growth spree and start proliferating? Are they capable of migration? And here’s the twist – when my protein of interest takes a dip, do other proteins decide to change their expression levels?
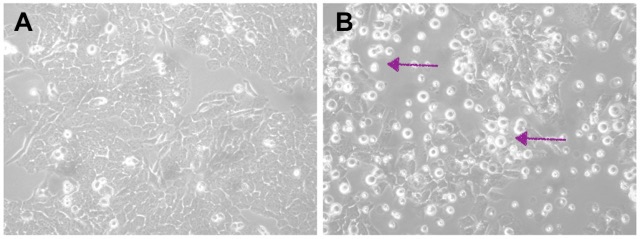
The picture below can probably help you get an idea of what I’ve done so far. Do you see those brighter spots in Pictures A and B? Those are dead cells. Their number indicates the proportion of dead cells after a treatment. Picture A has just a few; the majority are healthy and well-spread cells. This is our negative control, a condition when we show neuroblastoma cells that have been transfected, but no gene knockdown happened. Transfection is the term for introducing small RNA molecules. Now, in Picture B, when we knocked down the protein, it caused the death of the cells, and you can clearly see that from all those many little bright spots.
We have found answers to many of the previous questions, but new questions have arisen, and we can’t wait to answer them!
Written by Federica Cottone